Evolutionary Epidemiology – Lehtinen Group
We study the ecological and evolutionary mechanisms that shape the structure and diversity of bacterial populations, with a focus on the dynamics of public health relevant traits, such as antibiotic resistance. Our approach combines mathematical and statistical modelling with genomic and surveillance data. We are funded by an SNF PRIMA grant.
Strain structure and diversity
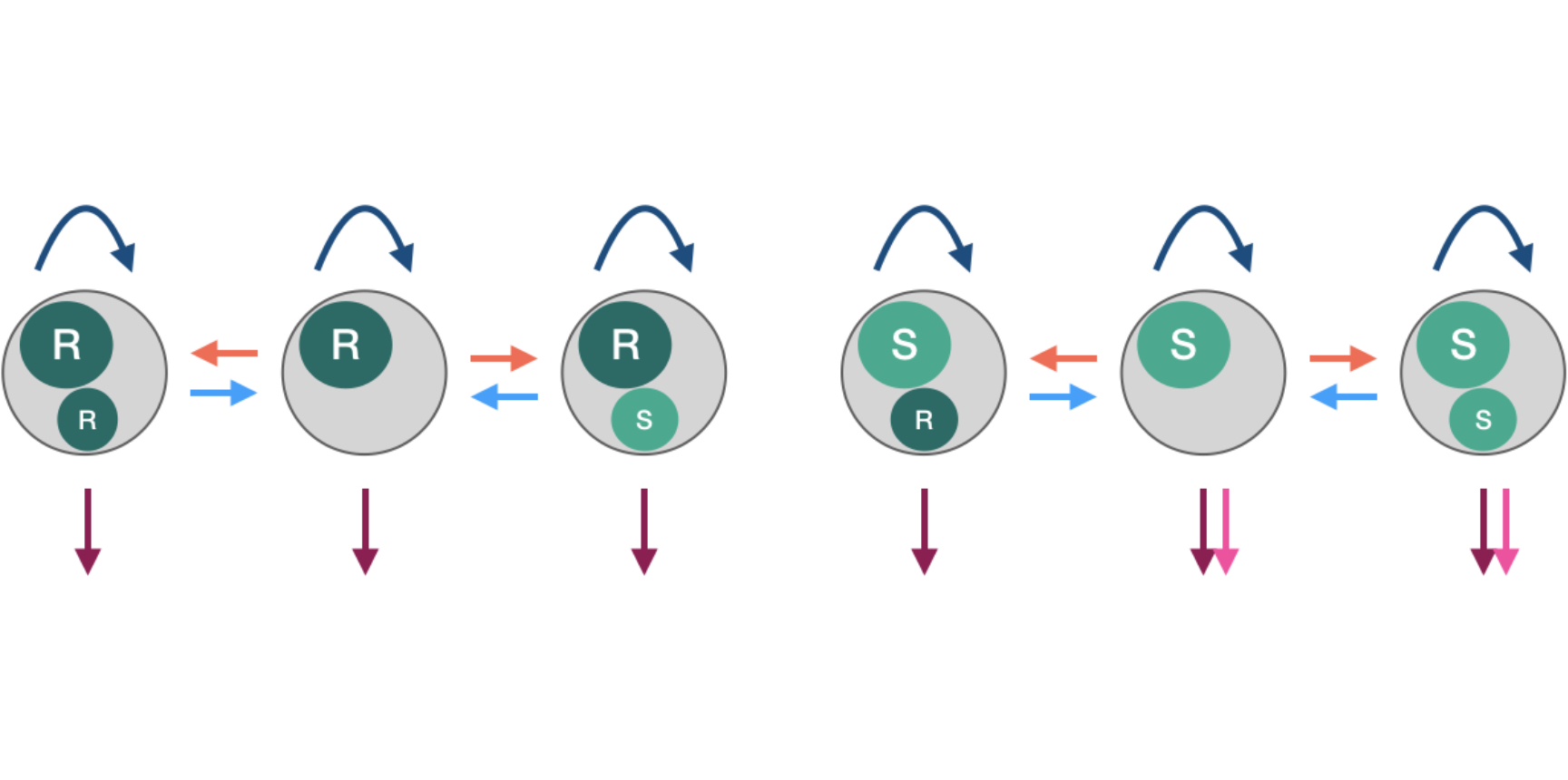
We are interested in a set of species that share a similar life-style (e.g. Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus and Streptococcus pneumoniae): generally commensal but occasionally pathogenic, and of high clinical relevance. These bacterial species exhibit considerable within-species diversity, with significant variation in important clinical traits (e.g. antibiotic resistance, immunological profile, virulence). Furthermore, these traits are not distributed randomly in the population: certain traits tend to co-occur, structuring the populations into distinct genotypes ('strain structure'). Our research tackles these two puzzles:
- what mechanisms explain persistent diversity despite competition for limited resources (i.e. hosts to infect)?
- what maintains strain structure despite high rates of gene transfer between strains?
Competition across scales
A key strand of our work is understanding how the interaction of within- and between-host scales shapes the competition between different strains (e.g. antibiotic sensitive and resistant strains). As part of this work, we are conducting a longitudinal stool sampling study in a nursery cohort to investigate strain dynamics within-host and transmission from host to host. We have also began developing some related theory on the effects of bacterial fratricide on strain dynamics at the epidemiological (i.e. host population) scale.
Plasmid ecology and evolution
Plasmids play an important role in bacterial evolution, in particular through their role as vehicles of horizontal gene transfer. Some genes, such as antibiotic resistance, are more likely to be found on plasmids than the chromosome. In recent work, we suggest a priority effect arising from positive frequency-dependent selection may explain this observation. In related work, we explore the effects of plasmid co-infection on the evolutionary and ecological dynamics of plasmids.